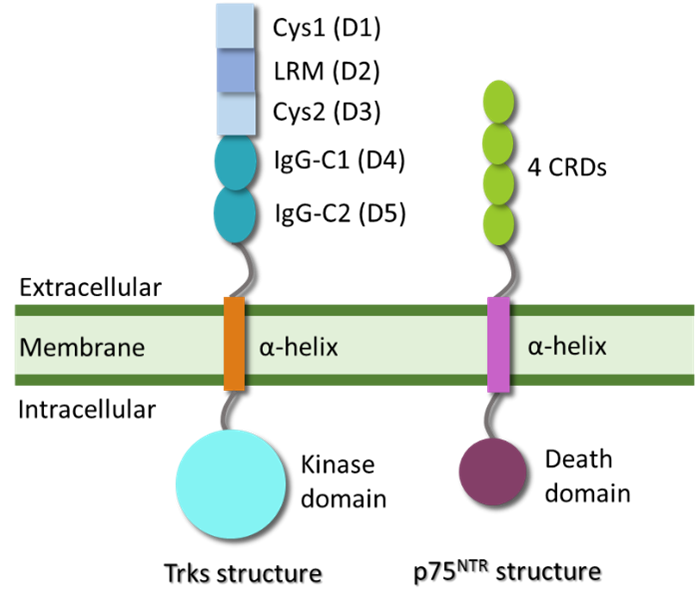
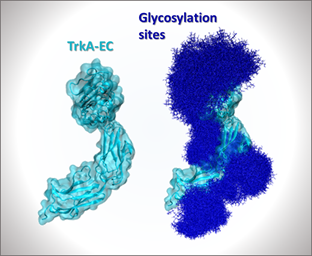
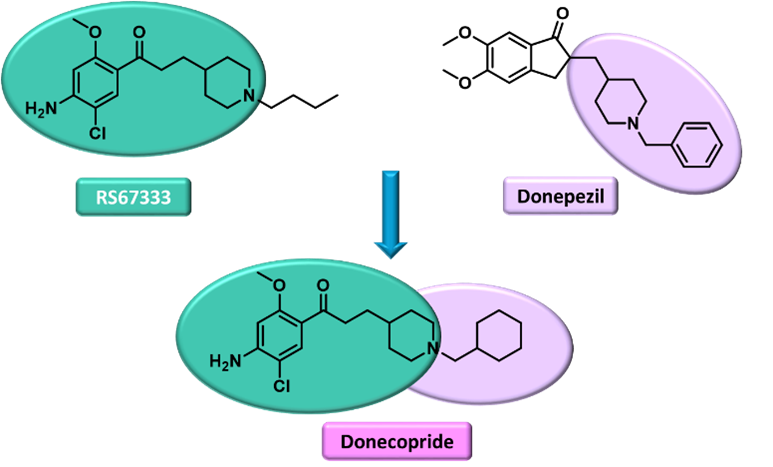
- Project
- Network
-
People
- Supervisors >
-
Early Stage Researchers
>
- ESR1: Daniele Narducci
- ESR2: Alessia Latorrata
- ESR3: Athanasios-Alexandros Tsengenes
- ESR4: Christina Athanasiou
- ESR5: Federica Carucci
- ESR6: Mirjana Antonijević
- ESR7: Paolo Giaccio
- ESR8: Canelif Yilmaz
- ESR9: Ana Aragón
- ESR10: Débora Pita
- ESR11: Thanasis Rogdakis
- ESR12: Desponia Charou
- ESR13: Evangelia Thanou
- ESR14: Marco Destro
- Scientific Advisory Board
- Training
- Publications
- Outreach
- News
- Job Openings
- Contact
- Project
- Network
-
People
- Supervisors >
-
Early Stage Researchers
>
- ESR1: Daniele Narducci
- ESR2: Alessia Latorrata
- ESR3: Athanasios-Alexandros Tsengenes
- ESR4: Christina Athanasiou
- ESR5: Federica Carucci
- ESR6: Mirjana Antonijević
- ESR7: Paolo Giaccio
- ESR8: Canelif Yilmaz
- ESR9: Ana Aragón
- ESR10: Débora Pita
- ESR11: Thanasis Rogdakis
- ESR12: Desponia Charou
- ESR13: Evangelia Thanou
- ESR14: Marco Destro
- Scientific Advisory Board
- Training
- Publications
- Outreach
- News
- Job Openings
- Contact
|
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 765704 |